View Winners →
View Winners → 
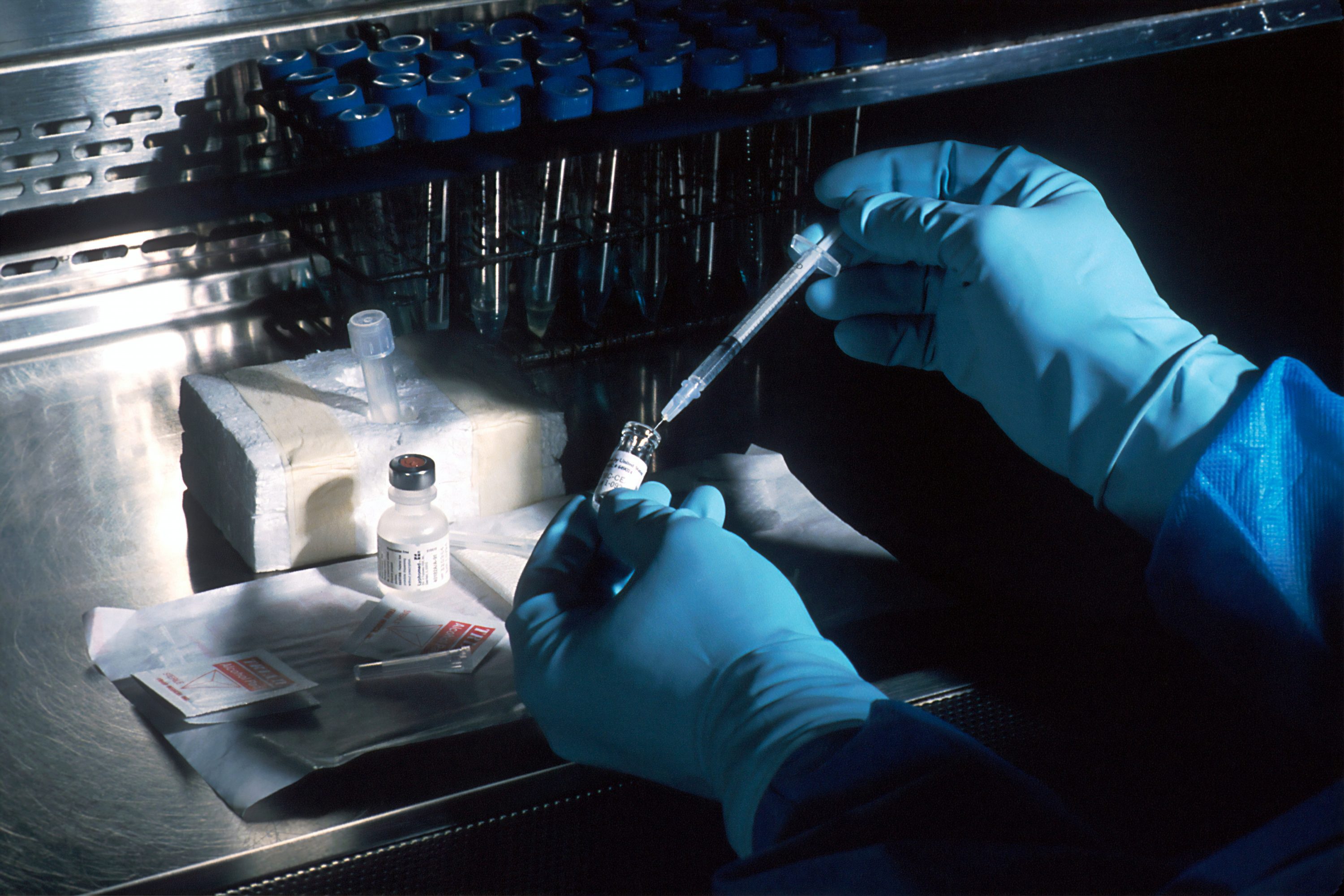
Photo by Medakit Ltd on Unsplash
UCSD researchers collaborated with colleagues from Baylor College of Medicine and Texas Children’s Hospital on a study involving experiments on flies to identify possible pathways to new drugs to treat COVID-19 and other viruses.
“A defining feature of viruses is their ability to rapidly evolve — a characteristic that has proven particularly challenging in controlling the SARS-CoV-2 virus,” said Ethan Bier, a professor with the UC San Diego School of Biological Sciences.
One approach to developing new treatments for such coronaviruses, including the SARS-CoV-2 virus that causes COVID-19, is to block the mechanisms by which the virus reprograms our cells and forces them to produce more viral particles. But studies have identified nearly 1,000 human proteins that have the potential to bind with viral proteins, creating overwhelming challenges in identifying which of the many possible interactions are most relevant to infection.
The multi-institutional collaboration has now developed a toolkit in fruit flies (Drosophila) to sort through the possibilities. The new Drosophila COVID Resource (DCR) provides a shortcut for assessing key SARS-CoV-2 genes and understanding how they interact with candidate human proteins.
“We envision that this new resource will offer researchers the ability to quickly assess the functional effects of factors produced by this once-in-a century pathogen as well as future naturally occurring variants.”
The researchers designed the DCR as a versatile discovery system. It features an array of fruit fly lines that produce each of the 29 known SARS-CoV-2 proteins and more than 230 of their key human targets. The resource also offers more than 300 fly strains for analyzing the function of counterparts to human viral targets.
“By harnessing the powerful genetic tools available in the fruit fly model system, we have created a large collection of reagents that will be freely available to all researchers,” said Hugo Bellen, a research team member from Baylor College of Medicine and Texas Children’s Hospital.
“We hope these tools will aid in the systematic global analysis of in vivo interactions between the SARS-CoV-2 virus and human cells at the molecular, tissue and organ level and help in the development of new therapeutic strategies to meet current and future health challenges that may arise from the SARS-CoV-2 virus and related family members.”
As they tested and analyzed the potential of the DCR, the researchers found that nine out of 10 SARS-CoV-2 proteins known as non-structural proteins (NSPs) they expressed in flies resulted in wing defects in adult flies. These defects can serve as a basis to understand how the viral proteins affect host proteins to disrupt or reorient essential cellular processes to benefit the virus.
They also made an intriguing observation: One of these viral proteins, known as NSP8, functions as a type of hub, coordinating with other NSPs in a mutually reinforcing manner. NSP8 also strongly interacted with five of the 24 human binding candidate proteins, the researchers noted. They discovered that the human protein that exhibited the strongest interactions with NSP8 was an enzyme known as arginyltransferase 1, or “ATE1.”
“ATE1 adds the amino acid arginine to other proteins to alter their functions,” said Annabel Guichard, a researcher from UC San Diego. “One such target of ATE1 is actin, a key cytoskeletal protein that is present in all of our cells.” Guichard noted that the researchers found much higher levels of arginine-modified actin than normal in fly cells when NSP8 and ATE1 were produced together. “Intriguingly, abnormal ring-like structures coated with actin formed in these fly cells,” Guichard added. “These were reminiscent of similar structures observed in human cells infected with the SARS-CoV-2 virus.”
However, when flies were given drugs that inhibit the activity of the human ATE1 enzyme, the effects of NSP8 were considerably reduced, offering a path to promising new therapeutics.
Calling their method a “fly-to-bedside” resource, the researchers say these initial results are just the tip of the iceberg for drug screening. Eight of the other NSPs they tested also produced distinctive phenotypes, laying the groundwork for pinpointing other new drug candidates.
“In several cases, identification of new candidate drugs targeting functionally important viral-human interactions might prove valuable in combination with existing anti-viral formulations such as Paxlovid,” said Bier. “These new discoveries may also provide clues to the causes of various long COVID symptoms and strategies for future treatments.”
Funding for the study was provided by the National Institutes of Health; the Kyoto Institute of Technology; the Tata Trusts in India to the Tata Institute for Genetics and Society at UC San Diego; Jan and Dan Duncan Neurological Research Institute at Texas Christian Hospital; and a CAPES fellowship.
The study was published in Cell Reports: www.cell.com/cell-reports/fulltext/S2211-1247(23)00853-7.